
In yeast and Arabidopsis, other proteins also promote crossover formation ( Figure 1). These results indicate that, in Arabidopsis, AtZYP1A and AtZYP1B are not ZMM proteins as defined in yeast. In addition, multivalents and bivalents between nonhomologous chromosomes are observed when both ATZYP1 genes are inactivated, showing that the AtZYP1 proteins are required to restrain the formation of crossovers between homologous chromosomes. Because no direct assay was carried out to study interference in this context, additional analysis is needed to confirm or to disprove the role of the AtZYP1 proteins in interference in Arabidopsis. Unlike the situation in yeast, in which the absence of Zip1 leads to a marked reduction in the number of crossovers, crossover formation is only slightly reduced (by 20%) when the two Arabidopsis orthologs of yeast ZIP1 ( ATZYP1A and ATZYP1B also known as ZYP1A and ZYP1B ) are inactivated using RNA interference. It had been proposed that the polymerization of the synaptonemal complex could be the interference signal, but there is now strong evidence against the idea of such a mechanism. Zip1, another ZMM protein in yeast, has a structural role in the synaptonemal complex. also estimated a proportion of $ 15% of noninterfering crossovers by fitting model data to genetic data. Therefore, the interfering pathway is responsible for the formation of most crossovers in Arabidopsis and involves AtMSH4 and AtMER3. The interference between the crossovers that are present is substantially reduced in atmer3 mutants. In Arabidopsis, the depletion of AtMSH4 and AtMER3 (also known as MER3), two of the ZMM proteins, greatly reduces the number of crossovers that form, with atmsh4 mutants having 85% fewer chiasmata than wild- type Arabidopsis and atmer3 mutants having 75% fewer. The actions of ZMM proteins indicate that interfering-crossover sites are selected from potential precursors at an early stage in the meiotic DSB-repair process. Noninterfering crossovers depend on the proteins Mus81 and Mms4. cerevisiae and Arabidopsis proteins are indicated here in parentheses: Rec107 (Mer2), Ski8 (Rec103), Mre2 (Nam8), Sae2 (Com1), Zip4 (Spo22), AtMSH5 (At3G20475), AtMUS81A (At4G30870) and AtMUS81B (At5G39770).
cerevisiae proteins but that have not been functionally characterized.
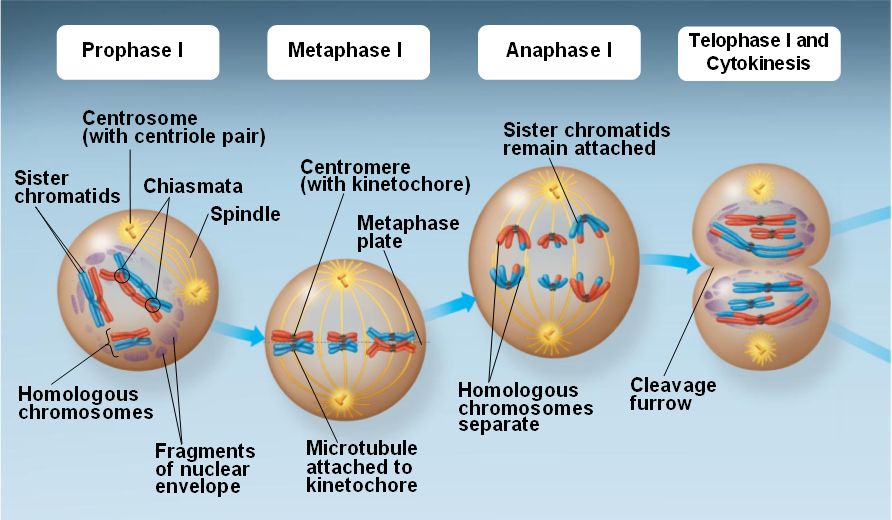
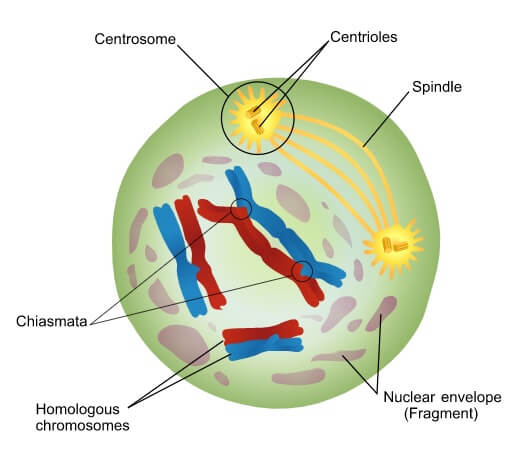
Question marks indicate Arabidopsis proteins that are thought to be involved in this process on the basis of their similarity with S. Proteins involved in this process are indicated, together with the stage they are thought to operate at data taken from Ref. Repair results in three different products: interfering crossovers, noninterfering crossovers and noncrossovers. DNA DSBs are formed at leptotene and are processed to form single strands, which then invade a chromatid of the homologous chromosome as a repair template. (b) A model of meiotic recombination pathways in Saccharomyces cerevisiae (yeast) and Arabidopsis thaliana. Then, at diakinesis (the final stage of prophase I), the synaptonemal complex has disappeared, and the further condensation of chromosomes reveals the presence of chiasmata. Next, at pachytene, synapsis is completed, and the synaptonemal complex links the homologs along their whole length. Then, at zygotene, the pairs of homologous chromosomes start to synapse, through formation of the synaptonemal complex. After replication, chromosomes condense into long threads. (a) The cytological progression of prophase I of meiosis.


 0 kommentar(er)
0 kommentar(er)
